A15 Tracing clonal evolution via transcriptome changes and somatic mutations
Cancers, including leukemias and lymphomas, do not present homogeneous cell populations, but often have complex subclonal structures. Moreover, upon therapy those subclonal structures can undergo significant shifts leading to a more aggressive and therapy-resistant tumor.
Here, we want to develop novel ways to reconstruct clonal evolution in non-solid tumors using population genetics and genomics approaches. We will develop methods to integrate various sources of information about clonal identity, with a special focus on leveraging information from gene expression profiles to resolve clonal genealogies in greater detail. The genealogies will then be used to make inferences about the tempo and mode of evolution in the clonal population.
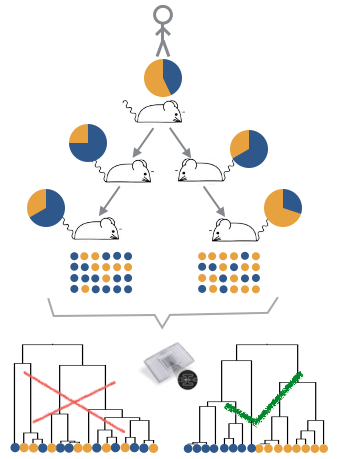
Figure legend: To evaluate how well we can reconstruct clonal genealogies from expression profiles, we will analyse single cell expression profiles and genotypes from patient derived AML-PDX mouse lines.We will generate 48 single cell expression profiles from 2 PDX mice each from 6 patient samples for which the presence of subclones has been confirmed via bulk genotyping. The subclones, should be identifiable by expressed somatic mutations, so that the subclonal type of each single cell can be determined from the RNA-Seq data. Then, we are going to reconstruct genealogies from the expression profiles, using different subsets of genes to correct for unwanted variance. We will choose a panel that best reflects the expected genealogy based on genotyping. Finally, we want to apply this analysis method in patient samples at diagnosis and relapse, we expect that the same panel also allows us to reconstruct the clonal genealogy of patients.


